Scripting in mricroGL for reproducible brain figures
mricroGL is a powerful program to produce high-resolution brain figures for posters and papaers. However the GUI is tricky to use and it doesn’t provide numerical feedback so it is difficult to produce the same slice of a brain using different masks.
One way to overcome this is to use scripting. The scripting terminal can be accessed by clicking on View - Scripting. Then you can input in some basic information through the following code. Be sure to replace the /PATH/TO/OVERLAY/OVERALY.nii.gz with an actual mask file.
Here is a code to produce a coronal slice looking at the striatum.
Coronal Slice
BEGIN
RESETDEFAULTS;
LOADIMAGE('mni152_2009bet');
BACKCOLOR(255,255,255);
OVERLAYLOADSMOOTH(TRUE);
OVERLAYLOAD('/PATH/TO/OVERLAY/OVERLAY.nii.gz');
OVERLAYCOLORNAME(1,'red_yellow');
OVERLAYMINMAX(1,0.25,1);
COLORBARVISIBLE(FALSE);
AZIMUTHELEVATION(0,0);
CUTOUT(0,0,0,1,0.62,1);
SHADERADJUST('overAlpha', 0);
END.
After running the script I usually slide overAlpha from the GUI to 0 to get rid of the masks that remain protruding the brain. I don’t think it’s an option in scripting yet*.
*Jeroen van Baar has notified me that you can actually also script the overAlpha by adding SHADERADJUST('overAlpha', 0);
The output would look something like this.
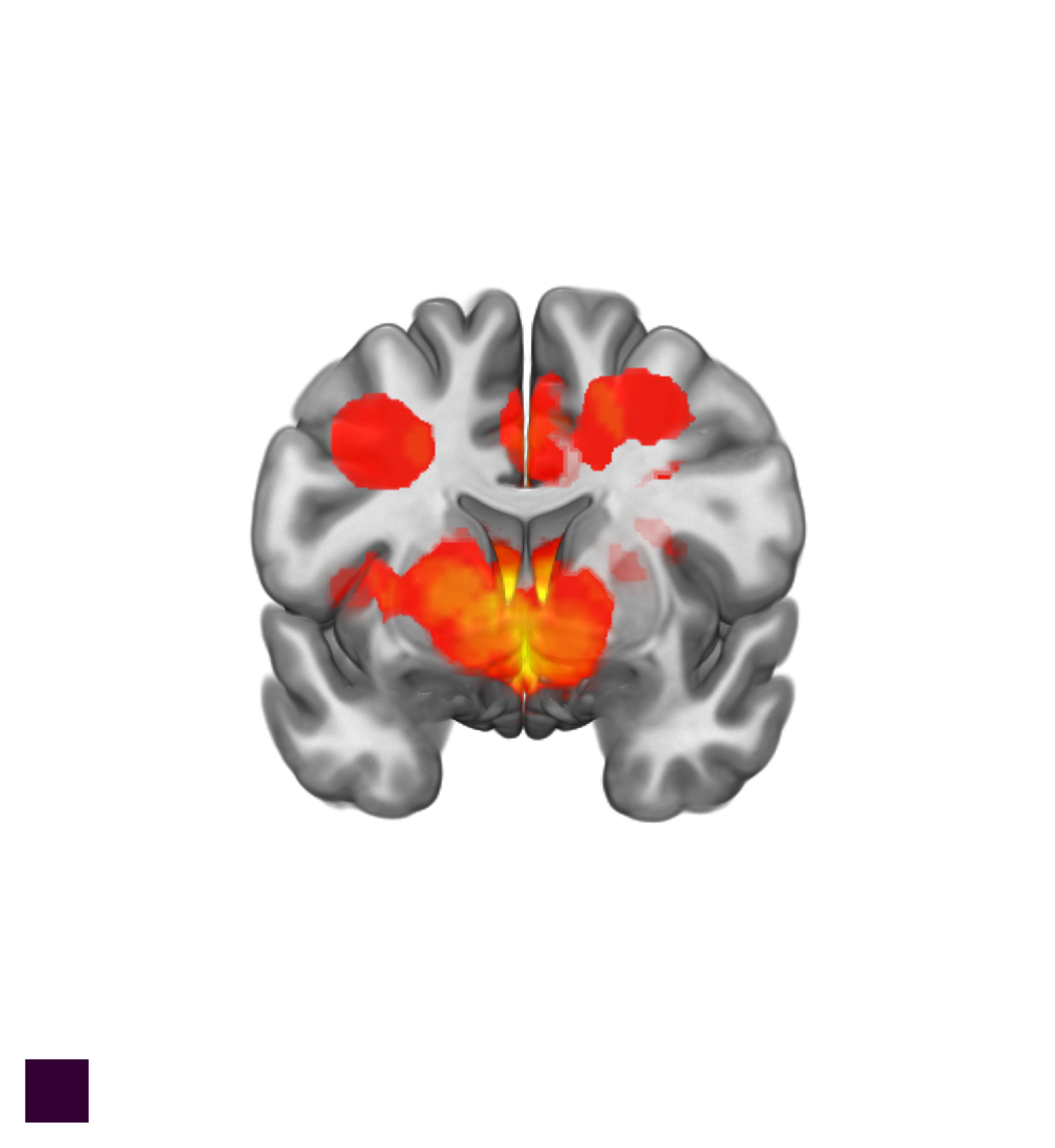
The AZIMUTHELEVATION controls the view angle and CUTOUT controls the slice.
The slice values are in ratios from the whole brain. Thus to change the view or slice you only need to modify the values for those two functions.
A lateral slice can be obtained by switching the last two lines with.
AZIMUTHELEVATION(0,90);
CUTOUT(0,0,1,1,1,0.4);
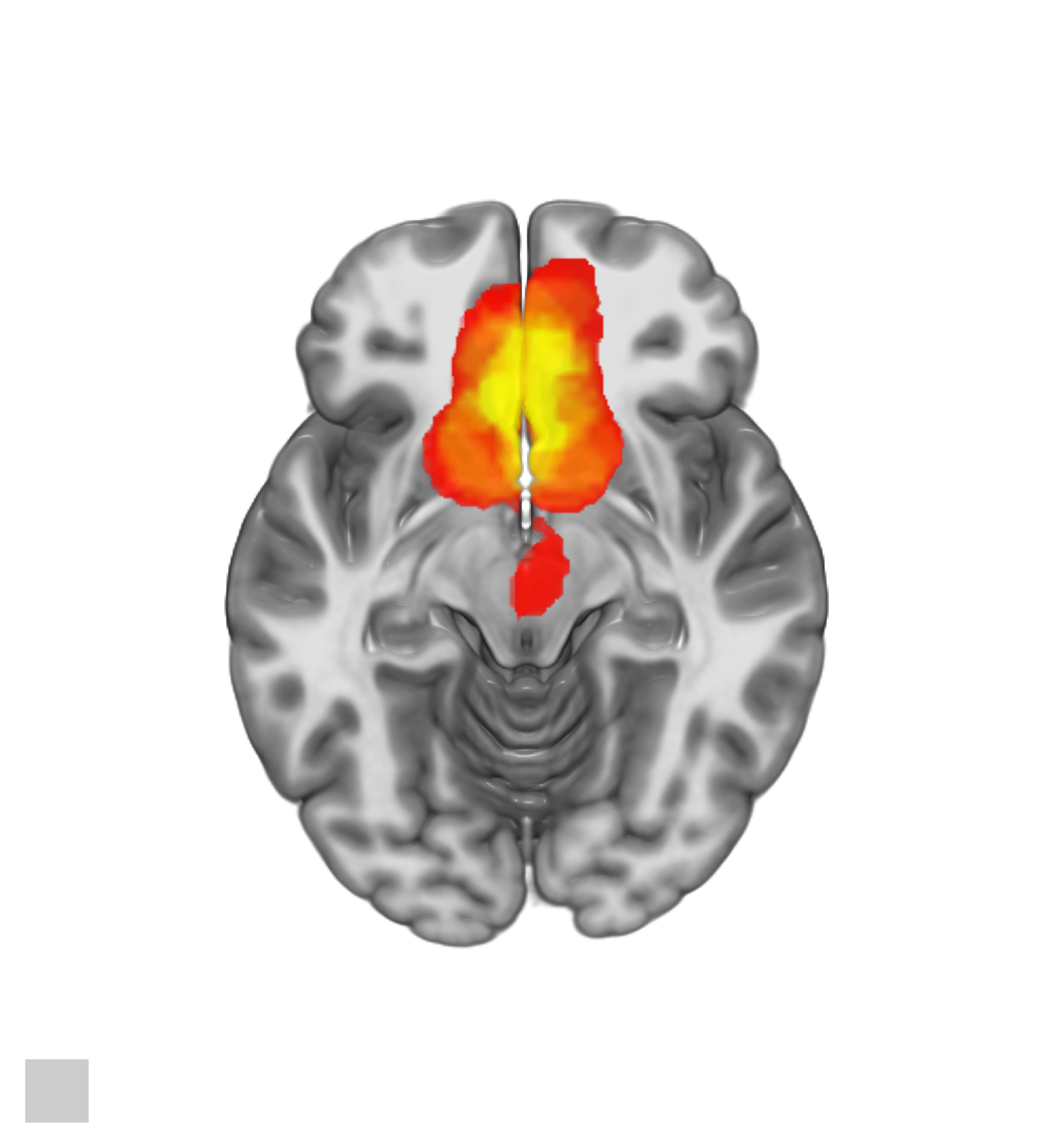
A saggital slice can be obtained by switching the last two lines with.
AZIMUTHELEVATION(-90,0);
CUTOUT(1,0,0,0.5,1,1);
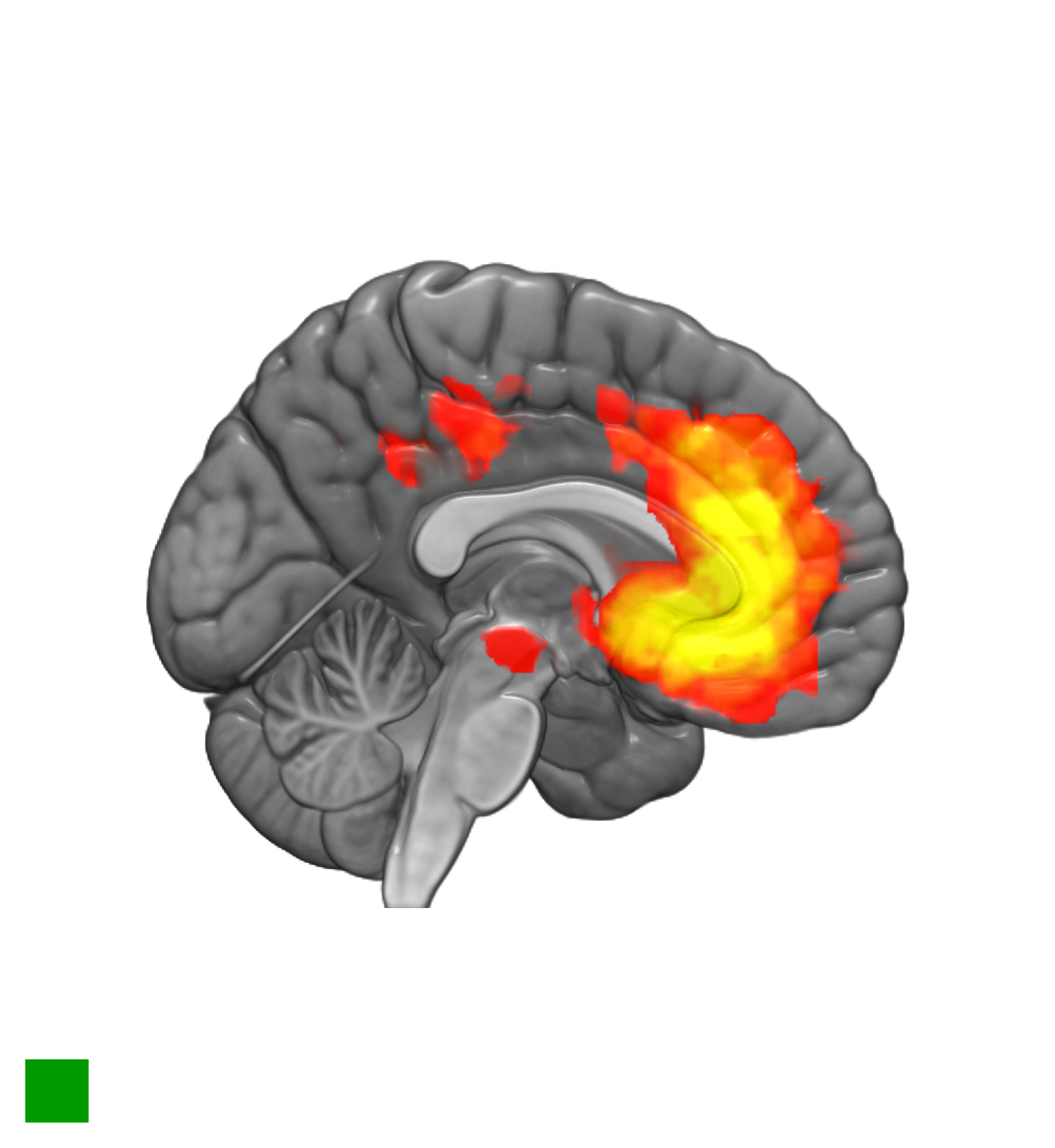
mricrogl fmri